Nano-phor
Welcome to the home page of the Nano-Phor research group. We are based both in the Avanced Biomedical Science and in the Physics Departments of the Federico II University of Naples, one of the largest and oldest research universities in Italy. Our activity is also carried out in the frame of the INFN (National Institute for Nuclear Physics) – Section of Naples.
Our research currently fucuses mostly on Bio-Nano-Photonics and the applications of Photonics to bio- and nano-sciences in a wide and multidisciplinary context. We design, fabricate and characterize novel materials at the nanoscale, using photonics characterization techniques but not only, dedicating a special attention to their applications to bio-sciences. We interpret this focus rather broadly, studying the interactions of novel materials such two-dimensional nanoflakes with biomolecules, bacteria, viruses, cells and soon also tissues. We combine these studies with the investigation of Ultrafast Molecular Dynamics of biologically relevant molecules such as nucleobases, proteins/antibodies and others, based on the ultrafast optical methods.
Among our interests we include also the nonlinear optical response of innovative materials and properly tailored surfaces, using methods typical of Nonlinear Optics. We also develop and employ theoretical and numerical methods to analyze genetic sequences and their corresponding transcripted peptides/proteins with the aim of studying their dynamics, their interactions and, for instance, predicting their mutations.
The group is also active in two collaborations with INFN, section Of Naples. Within the frame of the GINGER (Gyroscopes IN GEneral Relativity) collaboration, high-sensitivity interferometry based on gyroscopes is used with the goal to measure on the Earth the Lens-Thirring effect, predicted by General Relativity for the gravitational field of a rotating mass. Within the frame of the ARYA collaboration, our group is active in tailoring the morphology and the main properties of metallic surfaces, based on femtosecond laser ablation techniques, with the aim of minimizing secondary low-energy electron emission and realize the best cathodes for high-energy particle detectors.
advanced nonlinear And ultrafast optical spectroscopies
Supervisor
Carlo Altucci
Coordinator
Mohammadhassan Valadan
In our laboratory we use advanced nonlinear and ultrafast optical spectroscopic methods to study properties and dynamics of chemical reactions and molecular photo-dynamic in real time with a high temporal resolution of up to femtosecond time scale.
Time-resolved fluorescence measurements in nanosecond and femtosecond domains (“Time-Correlated Single Photon Counting” and “Fluorescence up-conversion” techniques, respectively) together with transient absorption spectroscopy in different spectral regions are employed to investigate, manipulate, and control ultrafast dynamics in complex materials. Our experiments span the mid infrared, visible, and ultraviolet regions of the spectrum to observe dynamics of vibrational and electronic state.
We develop and exploit such ultrafast multidimensional spectroscopic set-ups to unveil the most delicate details of processes in complex media, including biologically relevant molecules, nanostructures, and hybrid materials that are significantly desired for technological development and innovative applications.
With such capabilities, our research focuses on:
- Photo-dynamic properties of bio-molecules like 5-Benzyl-Uracil that is a model system for the cross-linking interaction between DNA and Proteins.
- Studying purposely designed and synthesized peptides to acquire information on cross-linking in biological samples when exposed to UV light.
- In a third type biologically relevant study, we monitor and control light-driven processes in engineered derivatives of Azo-compounds to exploit their application in drug delivery and activation by light.
Moreover, in a different context we use such intense ultrashort laser pulses to produce periodic micro- and nano- structures on different surfaces for applications like new surfaces for high energy photon detectors.
Two dimensional materials – Live matter surface interactions
Supervisor
Carlo Altucci
Coordinator
Manjot Singh
Imagine the beauty of science where the design and fabrication of a material opens up various other branches of science with very many practical applications and breakthrough inventions. Yes, it is true! This is how science works and this is how we all are connected with it and we gain multidisciplinary skills.
The two different aspects on which our group’s research activity is comprehended are: Green fabrication approach for different two dimensional materials (2DMs) such as graphene, graphene oxide, MoS2 and WS2 and their surface interaction studies with live matter such as biological cells and various microorganisms i.e. bacteria and viruses. In particular, we aim to expand our research activities in biomedical applications of 2DMs in use.
Brief Introduction to 2DMs
2DMs have gained a great deal of attention within the scientific community over the past decade owing to the unique new physics that emerges as we see transition from bulk to monolayer materials. In general, these are a group of ultrathin materials having 2D nanosheets (NSs) morphology with one or several atomic layers of thickness, large surface area combined with outstanding electronic, optical, electrochemical, mechanical and thermal properties. Because of these properties new channels for fundamental scientific research and advanced technological applications are opening, there including sensing, catalysis, various biomedical areas and functional nano-composites. Graphene, today’s wonder material is the first member of this 2DMs family for which in 2004, Nobel prize in Physics was awarded and after its discovery, it has sparked an intense research activity in very many application areas including its emerging commercialization.
Another important class of 2DMs is 2D transition metal dichalcogenides (TMDs) family. 2D TMDs have become the focus of fundamental research in various applications because of their outstanding physico-chemical properties, there including low cytotoxicity, tunable energy band gap and large surface area. Briefly, TMDs consist of a hexagonal layer of ransition metal atom (M) sandwiched between two layers of chalcogen atoms (X) with MX2 stoichiometry, where M ranges from Group IVB to Group VIII (Mo, W, Ti, V) and X represents the chalcogen (S, Se and Te).
Our contribution to this field / Our group’s research objective
- Green synthesis approach for 2DMs
Basically, environmental stability is one of the most important properties of a newly discovered nanomaterial which is targeted at large scale application. To unveil the important properties and full potential of 2D TMD NSs control over their production is very crucial.
So, our group focuses on the more versatile and scalable fabrication technique which is liquid phase exfoliation (LPE) using green solvents and functionalization with fungal protein such as hydrophobin to render them biocompatibility, long standing stability in the aqueous solution, additional surface charge and enhanced electrostatic interactions between different 2DMs. We have fabricated few layered bio functionalized MoS2, WS2 and graphene NSs using low boiling point solvents such as ethanol to study the long term stability and its implication in biosensing applications.
On the other hand, fabrication of 2DMs in pure water is a very challenging task, whereas it is highly essential to utilize these materials for core biomedical applications when prepared in physiological mediums. So, we have successfully fabricated 2D MoS2 in pure water using liquid phase exfoliation, having stability for one month which is very crucial for various biomedical tests. Suitable solvents, surfactants and other proteins can also be used to stabilize the nanosheets so as to avoid the reaggregation in the dispersion. Liquid processing of 2D nanosheets offer a wide range of materials to process and we have expanded our research activities to some new 2DMs such as antimonene which has excellent applications in biomedical field.
Suitable solvents, surfactants or polymers can be used to stabilize the nanosheets from reaggregation in the dispersion. So, we are also investigating a range of solvents, co-solvents considering their use in biomedical field. Furthermore, an extensive characterization of the fabricated 2D NSs using different tools such as UV-Visible and Raman spectroscopy, ζ-potential, AFM, SEM and TEM is also available to analyze the results.
- 2DMs – Live matter surface interaction studies
The main purpose of fabricating green technique based 2DMs is to understand their surface interactions with various live latter such as tumor and non-tumor cells, various microorganisms such as bacteria and viruses. It is highly important to know how these 2DMs behave at the very first surface membrane interaction so to understand their toxicological behavior on the live matter.
We have made a very interesting study on the biological interactions of 2D MoS2 nanosheets with tumor and non-tumor cells exhibiting a targeted anti-cancer effect on tumor cells whereas negligible effect on normal cells. This has paved a way for us to understand the mechanism behind this interesting study which we have been successful so far.
Similarly, we are also working on the anti-bacterial and ant-viral studies of 2D MoS2, garphene and graphene oxide on various pathogens such as two kinds of Salmonella typhimurium, E. coli and Staphylococcus aureus. These studies have shown very interesting results which we have proved mathematically in one of our recent studies.
Topological invariants and complex biomolecules
Supervisor
Carlo Altucci
Coordinator
Francesco Bajardi
Surprisingly enough, from very general and basic theories such as classical and quantum theories of gravity, ideas can lead to far beyond closely related fields, such as cosmology and astrophysics, to push concepts and applications to complex systems, there including the interactions between biomolecules, such as nucleic acids and proteins.
Despite its success, Einstein’s General Relativity (GR) presents many unsolved issues and puzzles at any scales. For instance, it is not able to predict the exponentially accelerated phases of the universe evolution without introducing the so called “Dark Energy”, or to describe the motion of the stars orbiting around the center of a given galaxy, without introducing the “Dark Matter”. Dark Energy and Dark matter are still unresolved rebus of the Universe and puzzling concepts to some extent. It is a fact that a complete and coherent theory capable together of describing the quantum world and explaining the cosmological and astrophysical observations is still missing; therefore, looking for such a theory is a very demanding research area and an exciting task.
While applications of modified theories of gravity to our universe seems to be a straightforward consequence for a testbed of the theory itself, the use of these theories in understanding complex systems might look unusual and non-conventional. Recently, the formalism adopted in many extended topological theories has been applied to describe several phenomena, mostly in astrophysics and cosmology. Topological field theories involves topological invariants to characterize the dynamics of the physical system; these latter are mathematical objects which does not depend on the local form of the spacetime, but only relies on its global structure, the topology. This means that once assigning the topology, a set of constant quantities arise, regardless of the curvature assumed by the spacetime point by point. For instance, most theories of gravity consider for the universe the topology of an expanding sphere, with exponential acceleration. Topological field theories have been successfully applied to the gravitational and the electromagnetic interactions, providing interesting results for theoretical physics. Another interesting application concerns biological systems that exhibit complicated topological structures, such as nucleic acids or proteins. This is particularly worth because of the complexity often assumed by biological systems. Different parts of the same molecule may assume a complicated three-dimensional shape, called tertiary structure. When two or more tertiary structures interact, the resulting system fold into a quaternary structure. The above mentioned nucleic acids (or proteins) are characterized by a tertiary and a quaternary structure. Schematization methods aimed at studying such structures represent one of the most controversial and discussed branch of science, due to the important implications in biology, microbiology, medicine etc. As an example, from the spatial configuration assumed by the DNA, it is possible to infer the place in which genomic mutations might occur, as well as the difference among phenotypes. Schematization methods can also help to provide the genetic (and epigenetic) probability to develop a certain disease. Another example is given by the interaction between cell DNA and virus DNA which, if well described, can lead to a comprehension of the corresponding infection evolution. From this point of view, the applications of Topological Field Theories to biological systems turn out to be powerful tools to investigate the docking among biological large molecules, or to simply schematize the DNA, its molecular configurations and its mutations. Again the link between GR-like theories and the dynamics/interactions of complex biomolecules is the topological nature of the former which can be essential to describe the complicated physico-chemical and biological behavior of the latter, very much relying on their topology. Basically, the main idea is to describe the DNA curvature by using the same formalism used for the spacetime, treating the interactions occurring in biological systems as driven by the same general principles that govern the gravitational interactions. As an example the figure below reports the curvature of a relevant biomolecule (Chern-Simons current value), namely receptor protein of the CD4 gene. Depending on the match between the curvature of CD4 and the curvature of the V3 glycoprotein present of the HIV virus membrane one can infer the likelihood of the virus attach to the host cell [1]
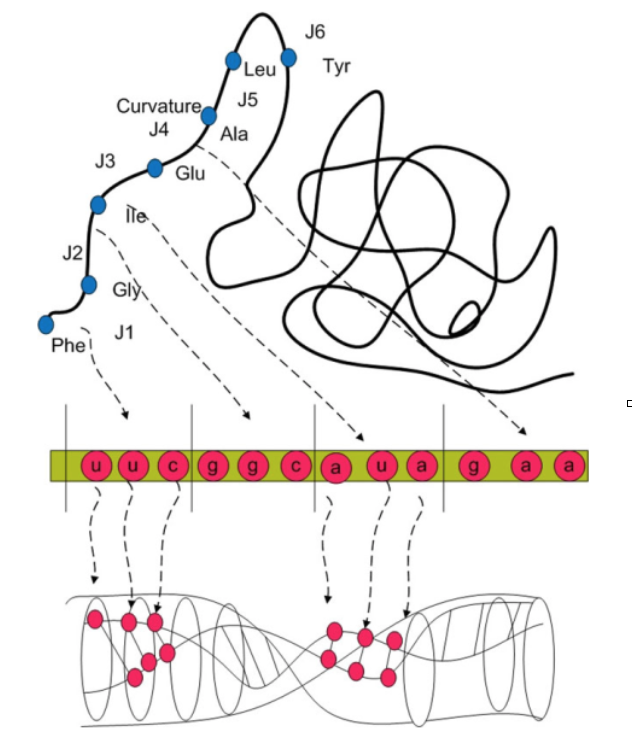
The picture shows the Chern-Simons current over CD4 receptor protein as curvature in behavior of docking states between V3 loop and CD4. The down panelis a loop space as Hopf fibration over DNA molecule.
Yet, bases/regions of the nucleic acids having the highest value of curvature might be the best candidates for a possible bond between molecules, similarly to compact and massive objects that cause the spacetime to be curved, thus attracting other bodies nearby. This approach is still under development and currently being applied to analyze a region of human KRAS gene and the evolution in space and time of the single-strand RNA CoVid virus sequence. The application consists in checking the curvature assumed by some selected part of the human gene KRAS, and comparing the original sequence with some mutated ones. The introduction of a mutation changes the point-like curvature of the given sequence and such difference may give important information regarding the biological impact of the corresponding mutated amino acid, as well as the frequency/probability of the mutation to occur. Another application currently under development is the analysis of COVID-19 virus sequences. The sequencing of more than 20K bases show that the mutations are focused on given preferred regions, depending on the country taken into account. The comparison among sequences of different countries might explain the reason why some countries are affected by a higher incidence of the virus during the 2020 pandemic.
In order to corroborate this method and to study other possible implications, also some standard modeling techniques are used. These techniques lie behind a probabilistic vision of the biomolecules interaction and make use of computational methods and software tools which aims to understand biological data. They combine different branches of science, such as mathematics, biology, computer science or statistics with the aim of better understanding the evolutionary aspects of molecular biology. The intrinsic probabilistic aspect of these techniques is necessary in order to treat systems made of a huge amount of particles. As the evolution of a particle can be well described by quantum or classical mechanics, many-bodies interactions are not such straightforward. Therefore, a statistical view of the biomolecule configurations is needed to infer the evolution of the system.
Merging the two different approaches (the one lying behind the above mentioned theories of gravity with the more conventional ones coming from bioinformatic), aims to implement the nowadays knowledge of the biological scenario. In particular, the deterministic aspect of the approach can be combined with bioinformatic approaches, which treat the biological issues from a probabilistic point of view by means of informatic methods. On the one hand, using topological field theories to describe DNA configuration can provide the exact position in which the mutation takes place by means of the comparison between the two sequence curvatures. Once the position of the mutation has been identified, bioinformatics is able to predict the probabilistic evolution and the clinical impact of that mutation. Moreover, even regarding the docking between macro-molecules, the probabilistic vision provided by bioinformatic techniques can be combined with the prediction given by topological field theories, in order to develop a coherent scheme capable of predicting where and when a disease could manifest. Bioinformatics, however, is a well defined and corroborated field and can be used for several other purposes. Some cases, in fact, cannot be treated under a topological view, such as linking regulatory DNA elements to their target genes, which may be located hundreds of kilobases away. With this purpose, bioinformatics uses an algorithm that identifies co-accessible pairs of DNA elements using single-cell chromatin accessibility data and so connects regulatory elements to their target genes. This algorithm, called Cicero, assembles groups of highly similar cells, using the clustering or pseudotemporal ordering.
PEOPLE
Former Members
Publications
Rusciano, G., Capaccio, A., Sasso, A., Singh, M., Valadan, M., Dell’Aversana, C., Altucci, L., Altucci, C.
(2022) Frontiers in Bioengineering and Biotechnology, 10, art. no. 844011
Molteni, E., Mattioli, G., Alippi, P., Avaldi, L., Bolognesi, P., Carlini, L., Vismarra, F., Wu, Y., Varillas, R.B., Nisoli, M., Singh, M., Valadan, M., Altucci, C., Richter, R., Sangalli, D.
(2021) Physical Chemistry Chemical Physics, 23 (47), pp. 26793-26805
DOI: 10.1039/d1cp04050b
Bajardi, F., Altucci, L., Benedetti, R., Capozziello, S., Sorbo, M.R.D., Franci, G., Altucci, C.
(2021) European Physical Journal Plus, 136 (10), art. no. 1080
Nivas, J.J.J., Valadan, M., Salvatore, M., Fittipaldi, R., Himmerlich, M., Rimoldi, M., Passarelli, A., Allahyari, E., Oscurato, S.L., Vecchione, A., Altucci, C., Amoruso, S., Andreone, A., Calatroni, S., Masullo, M.R.
(2021) Surfaces and Interfaces, 25, art. no. 101179
Di Virgilio, A.D., Altucci, C., Bajardi, F., Basti, A., Beverini, N., Capozziello, S., Carelli, G., Ciampini, D., Fuso, F., Giacomelli, U., Maccioni, E., Marsili, P., Ortolan, A., Porzio, A., Simonelli, A., Terreni, G., Velotta, R.
(2021) European Physical Journal C, 81 (5), art. no. 400
Capozziello, S., Altucci, C., Bajardi, F., Basti, A., Beverini, N., Carelli, G., Ciampini, D., Di Virgilio, A.D.V., Fuso, F., Giacomelli, U., Maccioni, E., Marsili, P., Ortolan, A., Porzio, A., Simonelli, A., Terreni, G., Velotta, R
(2021) European Physical Journal Plus, 136 (4), art. no. 394.
Benedetti, R., Bajardi, F., Capozziello, S., Carafa, V., Conte, M., Del Sorbo, M.R., Nebbioso, A., Singh, M., Stunnenberg, H.G., Valadan, M., Altucci, L., Altucci, C.
(2021) Analytical Letters, 54 (1-2), pp. 40-56.
Imperatore, C., Varriale, A., Rivieccio, E., Pennacchio, A., Staiano, M., D’auria, S., Casertano, M., Altucci, C., Valadan, M., Singh, M., Menna, M., Varra, M.
(2020) International Journal of Molecular Sciences, 21 (19), art. no. 7103, pp. 1-20.
DOI: 10.3390/ijms21197103
Singh, M., Zannella, C., Folliero, V., Di Girolamo, R., Bajardi, F., Chianese, A., Altucci, L., Damasco, A., Del Sorbo, M.R., Imperatore, C., Rossi, M., Valadan, M., Varra, M., Vergara, A., Franci, G., Galdiero, M., Altucci, C.
Combating Actions of Green 2D-Materials on Gram Positive and Negative Bacteria and Enveloped Viruses
(2020) Frontiers in Bioengineering and Biotechnology, 8, art. no. 569967, . Cited 3 times.
Allahyari, E., JJ Nivas, J., Avallone, G., Valadan, M., Singh, M., Granata, V., Cirillo, C., Vecchione, A., Bruzzese, R., Altucci, C., Amoruso, S.
(2020) Optics and Laser Technology, 126, art. no. 106073, . Cited 3 times.
Bosi, F., Di Virgilio, A.D.V., Giacomelli, U., Simonelli, A., Terreni, G., Basti, A., Beverini, N., Carelli, G., Ciampini, D., Fuso, F., Maccioni, E., Marsili, P., Stefani, F., Ortolan, A., Porzio, A., Altucci, C., Bajardi, F., Capozziello, S., Velotta, R.
(2020) Journal of Physics: Conference Series, 1468 (1), art. no. 012243, . Cited 1 time.
Imperatore, C., Valadan, M., Tartaglione, L., Persico, M., Ramunno, A., Menna, M., Casertano, M., Dell’aversano, C., Singh, M., Garigliota, M.L.D., Bajardi, F., Morelli, E., Fattorusso, C., Altucci, C., Varra, M.
(2020) International Journal of Molecular Sciences, 21 (4), art. no. 1246, . Cited 2 times.
DOI: 10.3390/ijms21041246
Virgilio, A.D.V.D., Belfi, J., Bosi, F., Giacomelli, U., Terreni, G., Beverini, N., Carelli, C., Ciampini, D., Ferrante, I., Fuso, F., Maccioni, E., Naletto, G., Ortolan, A., Altucci, C., Velotta, L., Porzio, A.
(2020) Journal of Physics: Conference Series, 1342 (1), art. no. 012084, .
Allahyari, E., JJ Nivas, J., Valadan, M., Fittipaldi, R., Vecchione, A., Parlato, L., Bruzzese, R., Altucci, C., Amoruso, S.
(2019) Applied Surface Science, 488, pp. 128-133. Cited 4 times.
Imperatore, C., Scuotto, M., Valadan, M., Rivieccio, E., Saide, A., Russo, A., Altucci, C., Menna, M., Ramunno, A., Mayol, L., Russo, G., Varra, M.
(2019) Journal of Photochemistry and Photobiology A: Chemistry, 377, pp. 109-118. Cited 3 times.
Valadan, M., Pomarico, E., Della Ventura, B., Gesuele, F., Velotta, R., Amoresano, A., Pinto, G., Chergui, M., Improta, R., Altucci, C.
(2019) Physical Chemistry Chemical Physics, 21 (48), pp. 26301-26310. Cited 2 times.
DOI: 10.1039/c9cp03839f
Beverini, N., Basti, A., Bosi, F., Carelli, G., Ciampini, D., Di Virgilio, A., Ferrante, I., Fuso, F., Giacomelli, U., Maccioni, E., Simonelli, A., Stefani, F., Terreni, G., Altucci, C., Porzio, A., Velotta, R.
(2019) Quantum Electronics, 49 (2), pp. 195-198. Cited 1 time.
DOI: 10.1070/QEL16881
Gesuele, F., JJ Nivas, J., Fittipaldi, R., Altucci, C., Bruzzese, R., Maddalena, P., Amoruso, S.
Analysis of nascent silicon phase-change gratings induced by femtosecond laser irradiation in vacuum
(2018) Scientific Reports, 8 (1), art. no. 12498, . Cited 15 times.
Kaur, J., Singh, M., Dell‘Aversana, C., Benedetti, R., Giardina, P., Rossi, M., Valadan, M., Vergara, A., Cutarelli, A., Montone, A.M.I., Altucci, L., Corrado, F., Nebbioso, A., Altucci, C.
(2018) Scientific Reports, 8 (1), art. no. 16386, . Cited 27 times.
Gesuele, F., Nivas, J.J.J., Fittipaldi, R., Altucci, C., Bruzzese, R., Maddalena, P., Amoruso, S.
(2018) Applied Physics A: Materials Science and Processing, 124 (2), art. no. 204, . Cited 3 times.
Nebbioso, A., Benedetti, R., Conte, M., Carafa, V., De Bellis, F., Shaik, J., Matarese, F., Della Ventura, B., Gesuele, F., Velotta, R., Martens, J.H.A., Stunnenberg, H.G., Altucci, C., Altucci, L.
(2017) Scientific Reports, 7 (1), art. no. 11725, . Cited 4 times.
Itri, F., Monti, D.M., Chino, M., Vinciguerra, R., Altucci, C., Lombardi, A., Piccoli, R., Birolo, L., Arciello, A.
(2017) Biochemical and Biophysical Research Communications, 492 (1), pp. 67-73. Cited 4 times.
Di Virgilio, A., Belfi, J., Bosi, F., Morsani, F., Terreni, G., Beverini, N., Carelli, G., Giacomelli, U., Maccioni, E., Ortolan, A., Porzio, A., Altucci, C., Velotta, R., Donazzan, A., Naletto, G., Cuccato, D., Beghi, A., Pelizzo, M.G., Ruggiero, M.L., Tartaglia, A., De Luca, G., Saccorotti, G.
(2017) Nuclear and Particle Physics Proceedings, 291-293, pp. 140-145. Cited 1 time.
Micciarelli, M., Curchod, B.F.E., Bonella, S., Altucci, C., Valadan, M., Rothlisberger, U., Tavernelli, I.
(2017) Journal of Physical Chemistry A, 121 (20), pp. 3909-3917. Cited 8 times.
Ventura, B.D., Iannaccone, M., Funari, R., Ciamarra, M.P., Altucci, C., Capparelli, R., Roperto, S., Velotta, R.
Effective antibodies immobilization and functionalized nanoparticles in a quartzcrystal microbalance-based immunosensor for the detection of parathion
(2017) PLoS ONE, 12 (2), art. no. e0171754, . Cited 26 times.
Kaur, J., Gravagnuolo, A.M., Maddalena, P., Altucci, C., Giardina, P., Gesuele, F.
(2017) RSC Advances, 7 (36), pp. 22400-22408. Cited 22 times.
DOI: 10.1039/c7ra01680h
Kaur, J., Vergara, A., Rossi, M., Gravagnuolo, A.M., Valadan, M., Corrado, F., Conte, M., Gesuele, F., Giardina, P., Altucci, C.
(2017) RSC Advances, 7 (79), pp. 50166-50175. Cited 11 times.
DOI: 10.1039/c7ra09878b
Porzio, A., Altucci, C., Capozziello, S., Velotta, R., Virgilio, A.D., Basti, A., Beverini, N., Carelli, G., Ciampini, D., Fuso, F., Giacomelli, U., Maccioni, E., Stefani, F., Terreni, G., Ortolan, A.
(2017) Proceedings of Science, 318, art. no. 181, .
DOI: link
Della Ventura, B., Ambrosio, A., Fierro, A., Funari, R., Gesuele, F., Maddalena, P., Mayer, D., Pica Ciamarra, M., Velotta, R., Altucci, C.
Simple and Flexible Model for Laser-Driven Antibody-Gold Surface Interactions: Functionalization and Sensing
(2016) ACS Applied Materials and Interfaces, 8 (33), pp. 21762-21769. Cited 4 times.
Funari, R., Della Ventura, B., Altucci, C., Offenhäusser, A., Mayer, D., Velotta, R.
(2016) Langmuir, 32 (32), pp. 8084-8091. Cited 20 times.
Belfi, J., Bosi, F., Di Virgilio, A., Beverini, N., Carelli, G., Giacomelli, U., Maccioni, E., Simonelli, A., Beghi, A., Cuccato, D., Donazzan, A., Naletto, G., Ortolan, A., Pelizzo, M.G., Porzio, A., Altucci, C., Velotta, R., Tartaglia, A.
(2016) 2016 European Frequency and Time Forum, EFTF 2016, art. no. 7477839, . Cited 3 times.
Della Ventura, B., Rea, I., Caliò, A., Giardina, P., Gravagnuolo, A.M., Funari, R., Altucci, C., Velotta, R., De Stefano, L.
(2016) Applied Surface Science, 364, pp. 201-207. Cited 9 times.
Itri, F., Monti, D.M., Della Ventura, B., Vinciguerra, R., Chino, M., Gesuele, F., Lombardi, A., Velotta, R., Altucci, C., Birolo, L., Piccoli, R., Arciello, A.
(2016) Cellular and Molecular Life Sciences, 73 (3), pp. 637-648. Cited 22 times.
Della Ventura, B., Funari, R., Anoop, K.K., Amoruso, S., Ausanio, G., Gesuele, F., Velotta, R., Altucci, C.
(2015) Applied Physics B: Lasers and Optics, 119 (3), pp. 497-501. Cited 3 times.
Funari, R., Della Ventura, B., Carrieri, R., Morra, L., Lahoz, E., Gesuele, F., Altucci, C., Velotta, R.
(2015) Biosensors and Bioelectronics, 67, pp. 224-229. Cited 62 times.
Valadan, M., D’Ambrosio, D., Gesuele, F., Velotta, R., Altucci, C.
(2015) Laser Physics Letters, 12 (2), art. no. 025302, . Cited 12 times.
Di Virgilio, A.D.V., Belfi, J., Bosi, F., Santagata, R., Beverini, N., Carelli, G., Maccioni, E., Simonelli, A., Ortolan, A., Altucci, C., Porzio, A., Velotta, R., Beghi, A., Cuccato, D., Donazzan, A., Naletto, G., Pelizzo, M.G., Tartaglia, A., Ruggiero, M.L., De Luca, G.
(2015) Proceedings of Science, 2-6-March-2015, art. no. 070, . Cited 1 time.
DOI: link
Funding
POR CAMPANIA FSE 2014-2020 ASSE III, 2022-2023, CUP B69J19000500007
INTERVENTI PER IL RAFFORZAMENTO DELL’OFFERTA REGIONALE DI INNOVAZIONE VERSO LE
IMPRESE DEL SETTORE AGRIFOOD
The Ministry of Education, University and Research (MIUR), 2019-2023 Project n. 20173B72NB
The Ministry of Education, University and Research (MIUR), PON (Programma Operativo Nazionale Ricerca e Innovazione) 2014-2020 (CCI 2014IT16M2OP005)









